Incomplete lineage sorting
Incomplete lineage sorting, also referred to as independent lineage sorting describes a phenomenon in population genetics when the tree produced by a single gene differs from the population or species level tree, producing a discordant tree. The ILS paradigm is characterized by segments of DNA that produce phylogenetic trees with different topologies compared to hypothetical inferred evolutionary trees.[1] In other words, in the species tree, the two closest relatives do not have the two most closely related alleles, instead they have more distantly related alleles. Incomplete lineage sorting can cause serious difficulties for phylogenetic inference.[2][3] Jeffrey Tomkins states:
| “ | Incomplete lineage sorting (ILS) is one of those perpetually confounding secrets in the field of biology that has plagued the fictional dogma of Darwinian evolution for many years.[4] | ” |
Examples
The Human GULO Pseudogene
The GULO gene (L-gulonolactone oxidase) has been detected in humans, apes, guinea pigs, bats, mice, rats, pigs, and passerine birds.[5] Loss of GULO gene function primarily impacts the cells ability to make vitamin C and has been detected among other animals in great apes and humans. Tomkins conducted a comparative analysis of the human and ape GULO genes and the results can be seen in the table below:
| The DNA sequence identity for each human GULO exon compared to five different ape taxon (DNA sequences extracted from the UCSC genome browser). Identities include insertions and deletions. | ||||||
|---|---|---|---|---|---|---|
| Percent Sequence Identity Compared to Human | ||||||
| Human GULO exon | Length (bases) | Chimp | Gorilla | Orangutan | Baboon | Rhesus |
| 1 | 101 | 99.1 | 100 | 97.1 | 91.1 | 91.1 |
| 2 | 96 | 98.0 | 98.0 | 94.8 | 92.8 | 88.6 |
| 3 | 107 | 98.2 | 85.0 | 98.2 | 74.8 | 74.8 |
| 4 | 151 | 99.4 | 90.8 | 97.4 | 94.1 | 92.8 |
| 5 | 164 | 97.6 | 97.0 | 94.6 | 91.5 | 90.3 |
| 6 | 129 | 97.7 | 98.5 | 99.3 | 95.4 | 93.8 |
| This Table: Author: Jeffrey P. Tomkins. Copyright owner: Answers in Genesis. Publication source: Answers Research Journal and its website, www.answersresearchjournal.org[5] | ||||||
According to Tomkins, the gorilla is considerably more similar to human in this region than chimpanzee—negating the inferred order of phylogeny. This pervasive evolutionarily discordant trend observed when constructing sequence-based trees, not only among humans and apes, but across the entire spectrum of eukaryotic life, is referred to as incomplete linage sorting.[5]
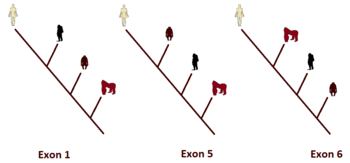
Looking at the DNA sequence identity table we can build a simple DNA similarity tree where we will find a discordant trend depending on which exon we use to build the tree. Using exon 1, the gorilla would be as close as possible to man; using exon 2, the gorilla and the chimpanzee would be tied; using exon 3, it is the chimpanzee and the orangutan that would be tied as the closest ape; using exon 4, the gorilla would be further away from baboons and Rhesus monkeys !; using exon 6, the orangutan would be the closest ape to the human being. Only exon 5 would bring the phylogeny expected by evolutionists. In the figure to the side, phylogenetic trees for exons 1, 5 and 6 are drawn for comparison.
The Bovini phylogeny and the Wisent
The wisent (Bison bonasus) or European bison and American bison are morphologically similar and cross-fertile and have closely related nuclear genes but the mtDNA phylogeny consistently clusters the wisent with the lineage leading to taurine cattle and zebu. A coalescent analysis indicated that the phylogeny discordance could be explained well by ILS alone according to Kun Wang et al.[6]
References
- ↑ Tomkins, Jeffrey; Bergman, Jerry (December 2013). "Incomplete lineage sorting and other ‘rogue’ data fell the tree of life". Journal of Creation 27 (3): 84–92. https://creation.com/rogue-data-fell-tree-of-life.
- ↑ Maddison, Wayne P; Knowles, L Lacey (February 2006). "Inferring Phylogeny Despite Incomplete Lineage Sorting". Systematic Biology 55 (1): 21–30.
- ↑ Futuyma, Douglas J. (2005). Evolution. Sunderland, Massachusetts: Sinauer. p. 38. ISBN 978-0-87893-187-3.
- ↑ Tomkins, Jeffrey (2012). More Than a Monkey: The Human-Chimp DNA Similarity Myth. CreateSpace Independent Publishing Platform. p. 72. ISBN 978-147525325-2.
- ↑ 5.0 5.1 5.2 Tomkins, Jeffrey (April 2, 2014). "The Human GULO Pseudogene—Evidence for Evolutionary Discontinuity and Genetic Entropy". https://answersingenesis.org/genetics/human-gulo-pseudogene-evidence-evolutionary-discontinuity-and-genetic-entropy/. Retrieved 2020-09-21.
- ↑ Wang, Kun; Lenstra, Johannes A.; Liu, Liang; Hu, Quanjun; Ma, Tao; Qiu, Qiang; Liu, Jianquan (19 October 2018). "Incomplete lineage sorting rather than hybridization explains the inconsistent phylogeny of the wisent". Communications Biology 1 (169).
External links
| |||||||||||||||||